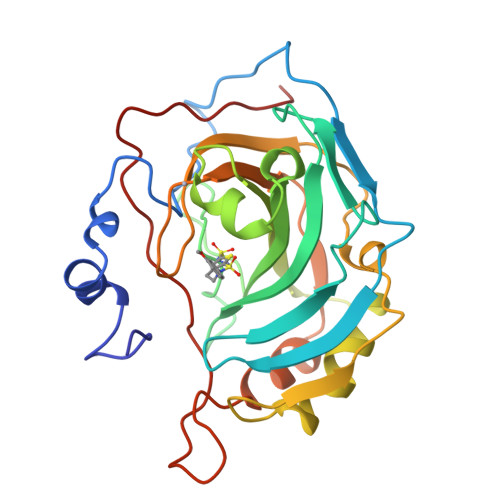
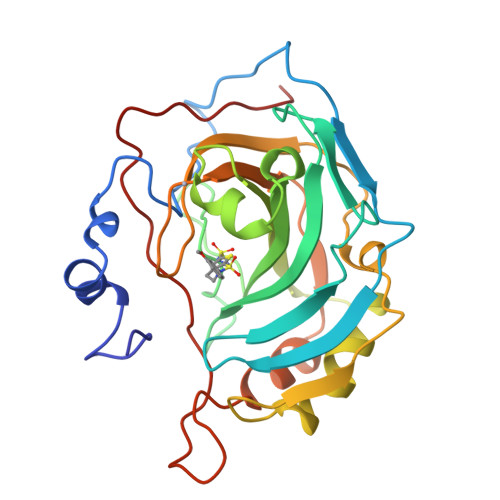
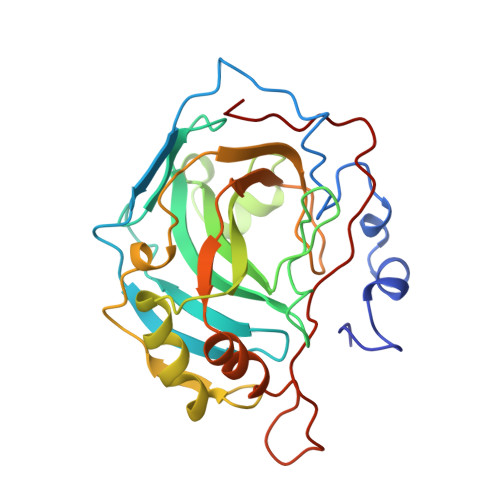
Structural analysis of inhibitor binding to human carbonic anhydrase II.
Boriack-Sjodin, P.A., Zeitlin, S., Chen, H.H., Crenshaw, L., Gross, S., Dantanarayana, A., Delgado, P., May, J.A., Dean, T., Christianson, D.W.(1998) Protein Sci 7: 2483-2489
- PubMed: 9865942
- DOI: https://doi.org/10.1002/pro.5560071201
- Primary Citation of Related Structures:
1BN1, 1BN3, 1BN4, 1BNM, 1BNN, 1BNQ, 1BNT, 1BNU, 1BNV, 1BNW - PubMed Abstract:
X-ray crystal structures of carbonic anhydrase II (CAII) complexed with sulfonamide inhibitors illuminate the structural determinants of high affinity binding in the nanomolar regime. The primary binding interaction is the coordination of a primary sulfonamide group to the active site zinc ion. Secondary interactions fine-tune tight binding in regions of the active site cavity >5 A away from zinc, and this work highlights three such features: (1) advantageous conformational restraints of a bicyclic thienothiazene-6-sulfonamide-1,1-dioxide inhibitor skeleton in comparison with a monocyclic 2,5-thiophenedisulfonamide skeleton; (2) optimal substituents attached to a secondary sulfonamide group targeted to interact with hydrophobic patches defined by Phe131, Leu198, and Pro202; and (3) optimal stereochemistry and configuration at the C-4 position of bicyclic thienothiazene-6-sulfonamides; the C-4 substituent can interact with His64, the catalytic proton shuttle. Structure-activity relationships rationalize affinity trends observed during the development of brinzolamide (Azopt), the newest carbonic anhydrase inhibitor approved for the treatment of glaucoma.
Organizational Affiliation:
Department of Chemistry, University of Pennsylvania, Philadelphia 19104-6323, USA.